Documentation
All of the key features in SlicerCineTrack will be available in the extension’s Inputs and Sequences section
- Open SlicerCineTrack by selecting the (1) dropdown menu > (2) SlicerCineTrack > (3) Track
- Load Inputs
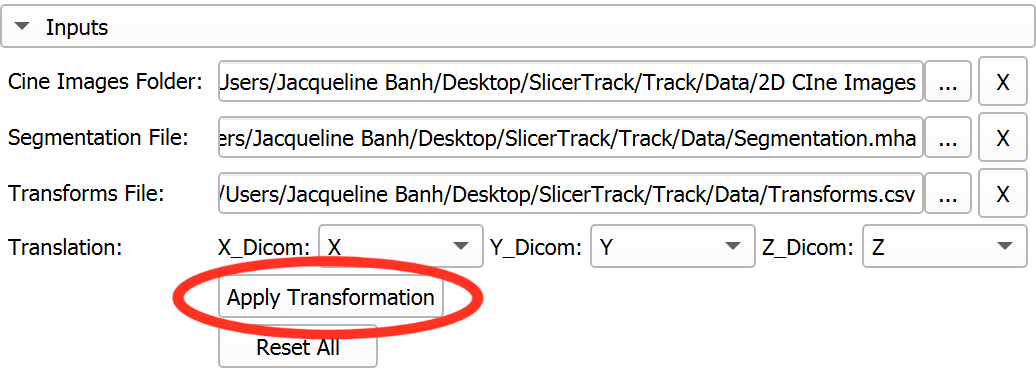
- Designate Transformation Columns - Translation:
- **X_Dicom:** Select the header for the X-direction transformations
- **Y_Dicom:** Select the header for the Y-direction transformations
- **Z_Dicom:** Select the header for the Z-direction transformations
- Click **Apply Transformations**
- Review Tracking Results
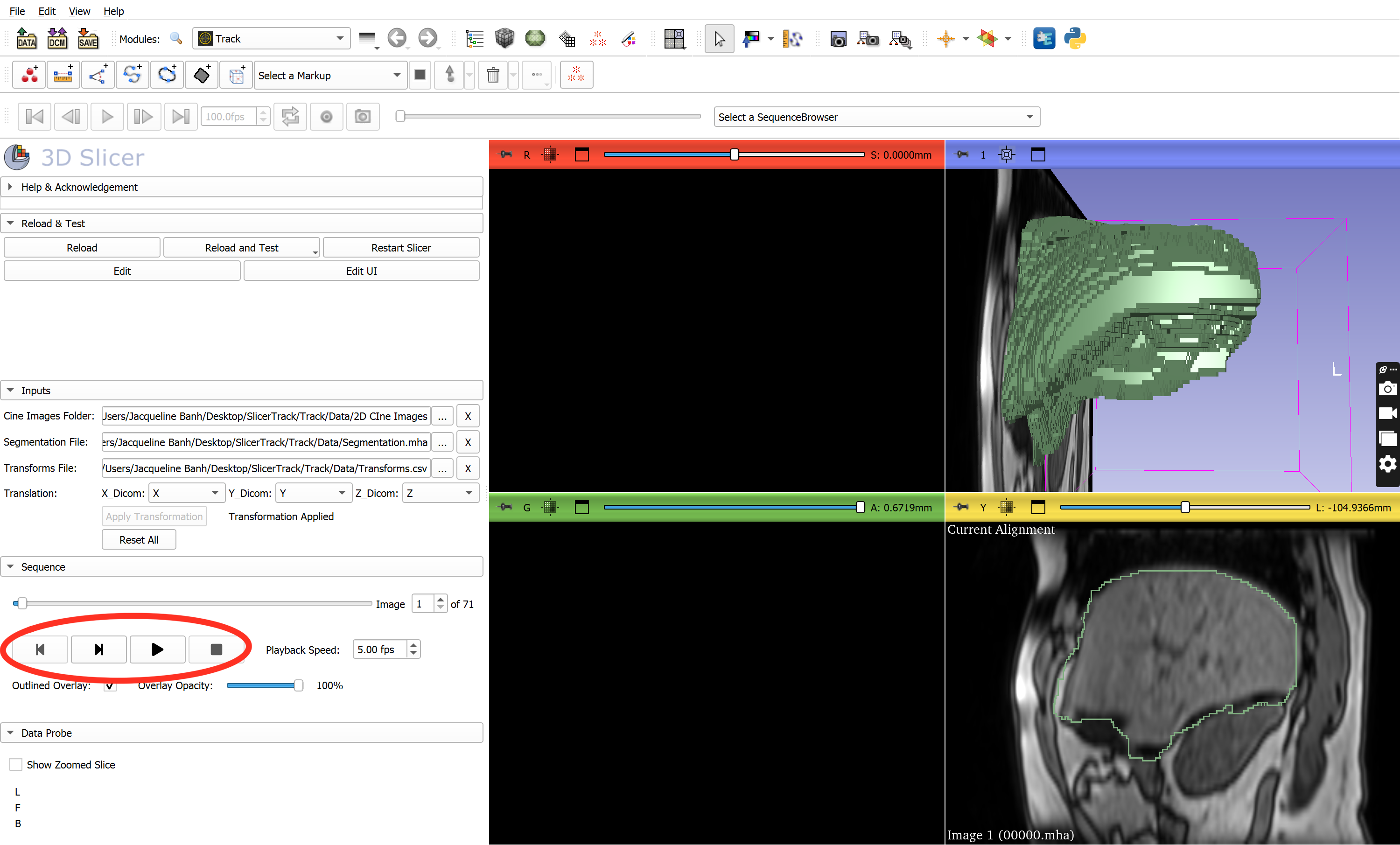
- After creating transform nodes from transformation data, the cine images and 3D segmentation are shown in the slice viewers
- Use the built-in toolbar to replay tracking data
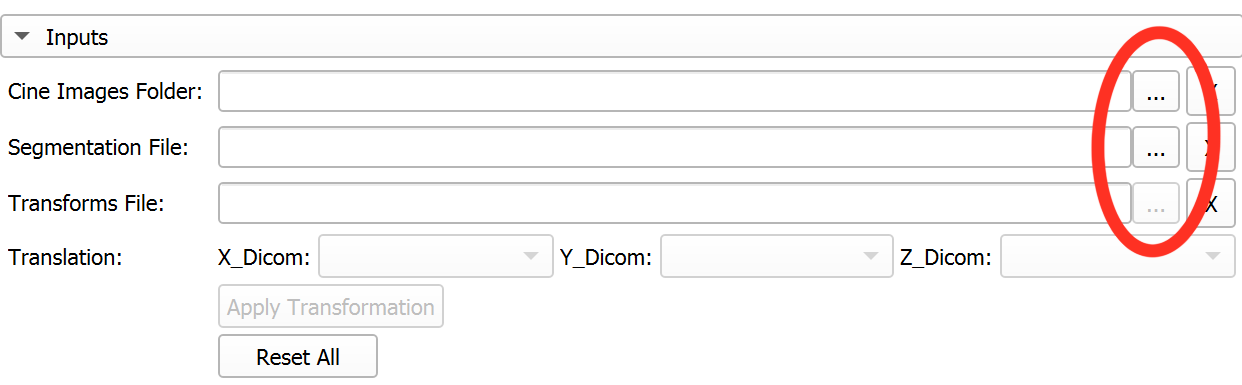
Click the button next to the file path textbox to select the inputs easily - **Cine Images Folder:** Select the folder with cine images - **Segmentation File:** Select the 3D segmentation file - **Transformation File:** Select a transformation file
Additional Features
- Play cine images zoomed in/out
- Adjust overlay opacity using the overlay opacity slider bar
- Use the image slider bar to move to a specific frame
- Insect frame number into the frame box to move to that specific frame
- Adjust the frames per second from 00.00 - 30.00 fps
- Click the X buttons to delete a specific input
- Click the Reset All button to remove all inputs
Sample Dataset
Dear Users,
This dataset was created based on a dataset sourced from the Cancer Imaging Archive (TCIA), and more specifically:
Hugo, G. D., Weiss, E., Sleeman, W. C., Balik, S., Keall, P. J., Lu, J., & Williamson, J. F. (2016). Data from 4D Lung Imaging of NSCLC Patients (Version 2) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2016.ELN8YGLE
Before accessing or utilizing this data, please refer to and adhere to the TCIA data use policy.
Warm regards
Sample Data set can be download from here